 Help DocsStrobe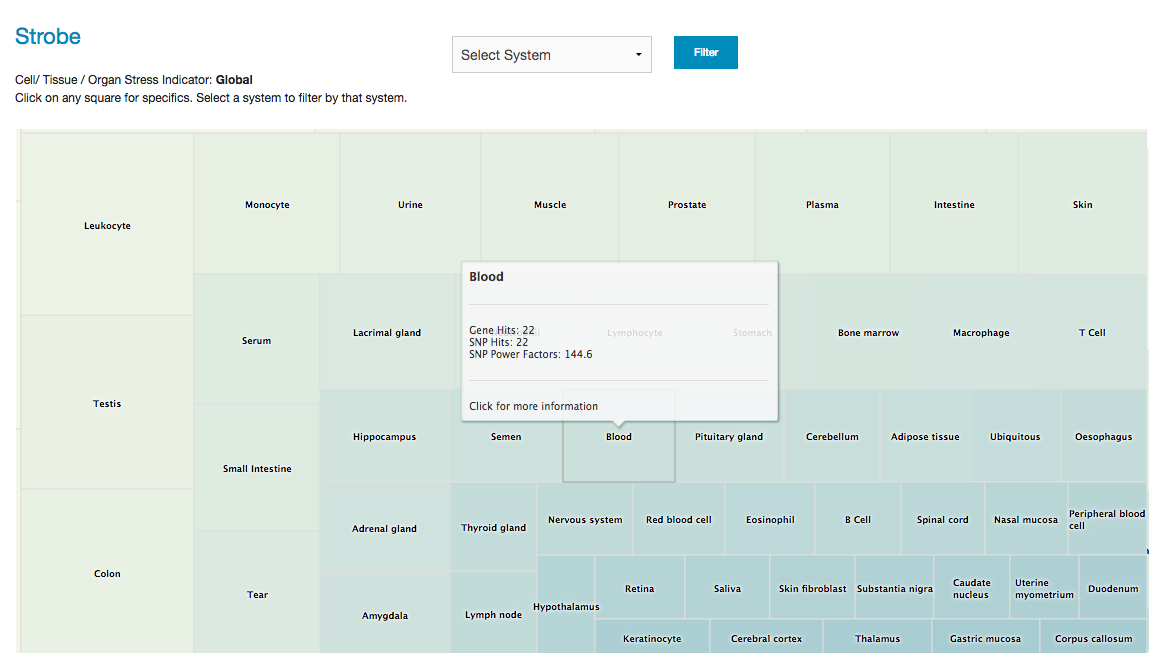
The STROBE app provides information on the influence of genetic variation on specific body tissues. Opus23 Pro uses a mashup of internal SNP data with gene tissue expression data derived from a variety of public databases to produce a comparative indicator of potential genetic stressors on body tissues arranged by system. Getting around STROBE From the 'Informatics' link on the main menu, hover over 'Analytics' until a second menu list appears, then select ‘Strobe’. You will then be presented with the default global STROBE screen as a squarified heat-map. The organ or tissue with the highest values have a lighter color, and are arranged from the highest value in the upper left corner to lowest value the bottom right corner. Values are assigned as an aggregate of power factors of client SNP mutations and the number of genes associated with that tissue. To filter from the default global view you can use the pull-down menu to limit tissues to specific systems, just click on Filter after selecting the system. Return to the default view by clicking on the Global View button. Here we limit the display to tissues, organs and cells of the immune system: 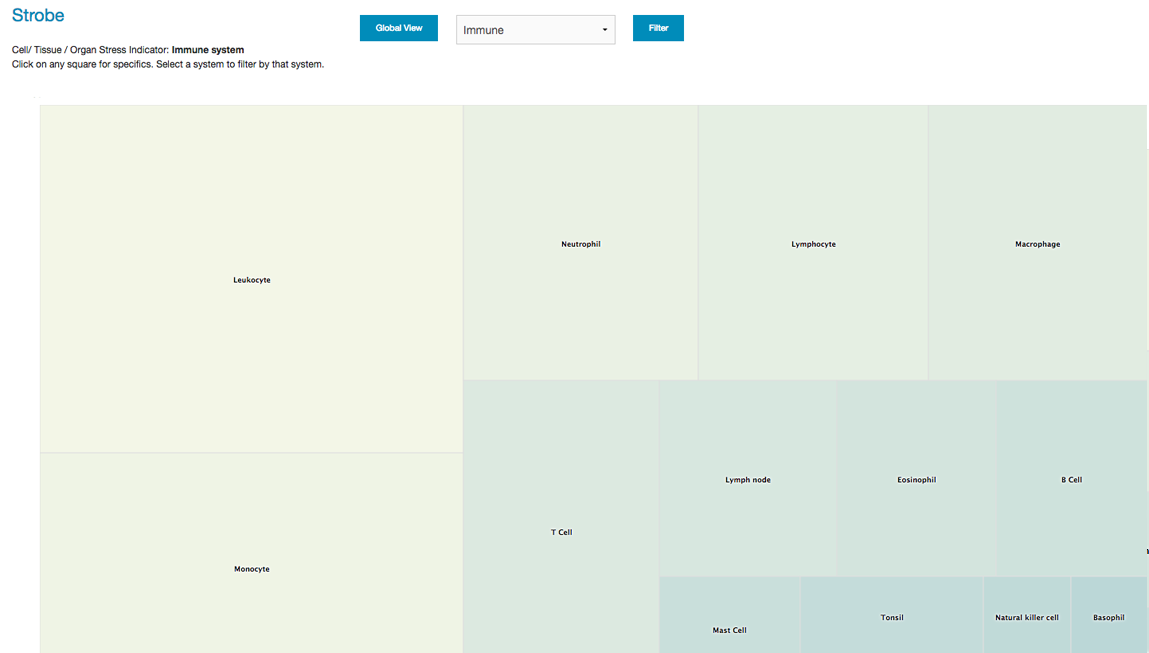
Clicking on any tissue brings up a modal popup with a Manhattan-type gene distribution display. The Y axis shows cumulative power factors of SNPs in that gene with risk variations. The X axis is a rough approximation of gene locus position. Yellow points indicate genes with exclusively heterozygous mutations. Orange points denote genes that contain homozygous mutations. Drag a rectangle around any area to zoom into that part of the map. Use the ‘Reset Zoom’ to return to full zoom. 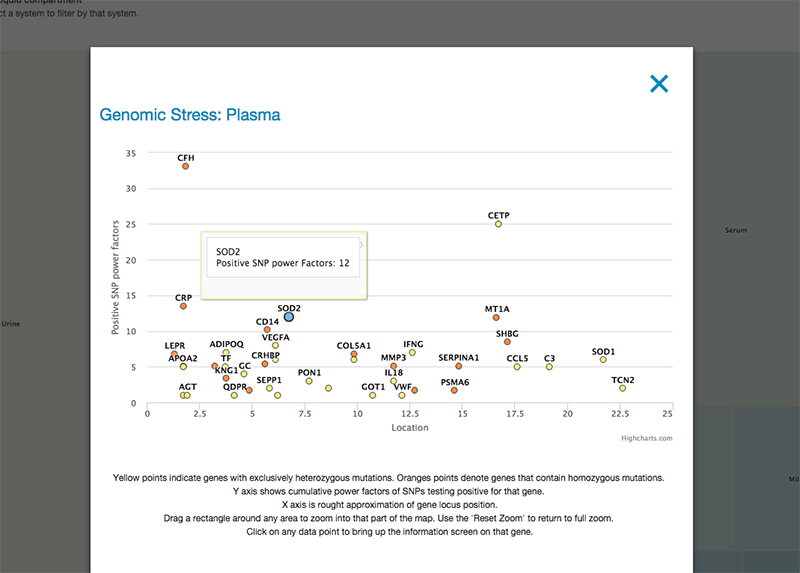
Clicking on any gene data point in the pop-up brings up the information popup for that gene. From there you can notate, or investigate associated agents, SNPs and algorithms. |
